知识图谱构建
构建概述
设计基于neo4j的中医知识图谱的构建方案,为智能诊疗系统提供结构化的医学知识支撑,虽然当前版本主要依赖GLM-4.1V的内在知识,但为未来扩展奠定基础。(仍在开发中)
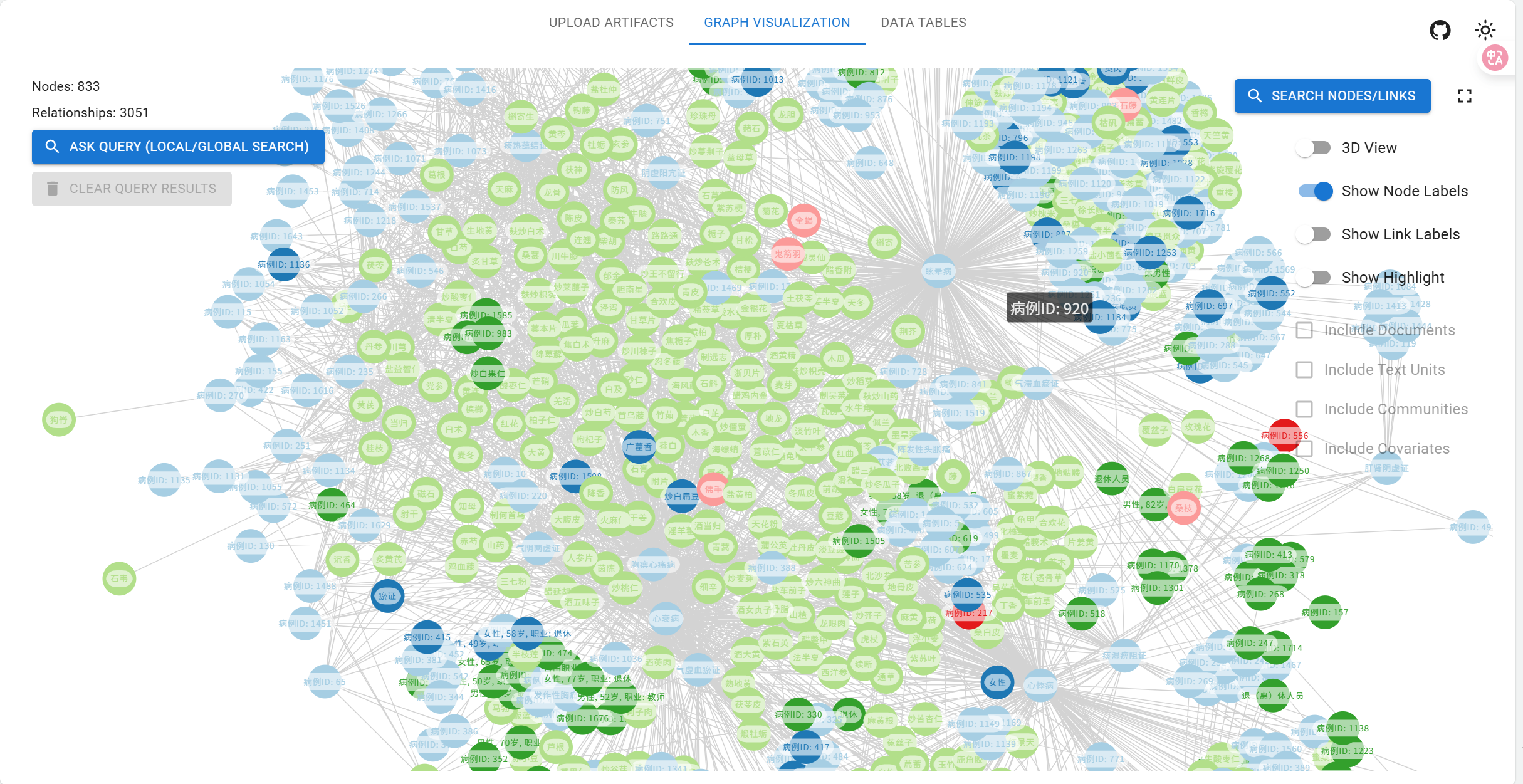
中医专病知识图谱示例
知识图谱设计架构
图谱整体结构
中医知识图谱主要包含以下核心实体和关系:
核心实体类型:
- 疾病实体:脾胃病、胃痛、腹胀、消化不良等
- 症状实体:胃脘疼痛、食欲不振、腹部胀满等
- 证候实体:脾胃虚弱、胃热炽盛、寒凝胃脘等
- 治法实体:健脾益气、清热和胃、温中散寒等
- 方剂实体:四君子汤、保和丸、理中汤等
- 药物实体:人参、白术、陈皮、山楂等
关系类型设计:
python
relationship_types = {
# 症状-疾病关系
"symptom_of": "症状属于某疾病",
"main_symptom": "主要症状",
"accompanying_symptom": "伴随症状",
# 证候-疾病关系
"syndrome_of": "证候属于某疾病",
"differential_diagnosis": "鉴别诊断",
# 治疗关系
"treats": "治疗关系",
"formula_for": "方剂适应症",
"herb_in_formula": "药物组成",
# 因果关系
"causes": "导致关系",
"aggravates": "加重关系",
"relieves": "缓解关系"
}实体识别与抽取
中医实体识别
基于规则的实体识别:
python
class TCMEntityRecognizer:
def __init__(self):
self.disease_patterns = [
r'(脾胃|胃脘|腹部).{0,3}(疼痛|胀满|不适)',
r'(消化|食欲).{0,3}(不良|不振|减退)',
r'(胃|脾).{0,2}(虚|实|热|寒)'
]
self.symptom_patterns = [
r'(胃脘|上腹|脐周).{0,3}(疼痛|胀痛|隐痛)',
r'(食后|饭后|进食后).{0,3}(胀满|不适|疼痛)',
r'(大便|便).{0,2}(溏|干|秘|稀)'
]
self.herb_patterns = [
r'(人参|党参|太子参)',
r'(白术|苍术|炒白术)',
r'(陈皮|橘皮|新会陈皮)'
]
def extract_entities(self, text):
"""
从文本中提取中医实体
"""
entities = {
'diseases': [],
'symptoms': [],
'herbs': [],
'syndromes': []
}
# 疾病实体识别
for pattern in self.disease_patterns:
matches = re.findall(pattern, text)
entities['diseases'].extend(matches)
# 症状实体识别
for pattern in self.symptom_patterns:
matches = re.findall(pattern, text)
entities['symptoms'].extend(matches)
# 中药实体识别
for pattern in self.herb_patterns:
matches = re.findall(pattern, text)
entities['herbs'].extend(matches)
return entities基于深度学习的实体识别:
python
class NERModel:
def __init__(self, model_path):
self.model = AutoModelForTokenClassification.from_pretrained(model_path)
self.tokenizer = AutoTokenizer.from_pretrained(model_path)
self.label_map = {
'B-DISEASE': '疾病开始',
'I-DISEASE': '疾病延续',
'B-SYMPTOM': '症状开始',
'I-SYMPTOM': '症状延续',
'B-HERB': '中药开始',
'I-HERB': '中药延续',
'O': '非实体'
}
def predict_entities(self, text):
"""
使用深度学习模型识别实体
"""
# 文本编码
inputs = self.tokenizer(text, return_tensors="pt", truncation=True, padding=True)
# 模型推理
with torch.no_grad():
outputs = self.model(**inputs)
predictions = torch.argmax(outputs.logits, dim=-1)
# 解码实体
tokens = self.tokenizer.convert_ids_to_tokens(inputs["input_ids"][0])
labels = [self.label_map.get(pred.item(), 'O') for pred in predictions[0]]
# 提取实体
entities = self.extract_entities_from_labels(tokens, labels)
return entities关系抽取与构建
症状-疾病关系抽取
模式匹配方法:
python
class SymptomDiseaseRelationExtractor:
def __init__(self):
self.relation_patterns = [
{
'pattern': r'(.+?)(?:常见于|多见于|是)(.+?)(?:的|之)(?:主要|常见)?症状',
'relation': 'symptom_of',
'entities': ['symptom', 'disease']
},
{
'pattern': r'(.+?)(?:患者|病人)(?:常|多|易)(?:出现|发生)(.+?)',
'relation': 'has_symptom',
'entities': ['disease', 'symptom']
}
]
def extract_relations(self, text):
"""
提取症状-疾病关系
"""
relations = []
for pattern_info in self.relation_patterns:
matches = re.findall(pattern_info['pattern'], text)
for match in matches:
relation = {
'relation_type': pattern_info['relation'],
'head_entity': match[0].strip(),
'tail_entity': match[1].strip(),
'confidence': 0.8
}
relations.append(relation)
return relations治疗关系构建
方药关系抽取:
python
class TreatmentRelationBuilder:
def __init__(self):
self.formula_herb_patterns = [
r'(.+?汤|.+?丸|.+?散)(?:由|含|包括)(.+?)(?:组成|构成)',
r'(.+?)(?:方|剂).*?(?:用|取|以)(.+?)(?:为|作为)(?:主|君|臣)药'
]
self.treatment_patterns = [
r'(?:治疗|主治|用于)(.+?)(?:可|宜|应)(?:用|服|投)(.+?)(?:汤|丸|散)',
r'(.+?)(?:证|症).*?(?:治|方|药)(?:以|用|宜)(.+?)'
]
def build_treatment_relations(self, medical_texts):
"""
构建治疗关系
"""
treatment_graph = {
'formula_herb': [], # 方药组成关系
'syndrome_treatment': [], # 证候治疗关系
'herb_efficacy': [] # 药物功效关系
}
for text in medical_texts:
# 提取方药组成关系
formula_relations = self.extract_formula_relations(text)
treatment_graph['formula_herb'].extend(formula_relations)
# 提取治疗关系
syndrome_relations = self.extract_syndrome_relations(text)
treatment_graph['syndrome_treatment'].extend(syndrome_relations)
return treatment_graph知识融合与验证
多源知识融合
知识源整合:
python
class KnowledgeIntegrator:
def __init__(self):
self.knowledge_sources = {
'classical_texts': ClassicalTextProcessor(), # 经典文献
'clinical_cases': ClinicalCaseProcessor(), # 临床案例
'modern_research': ResearchPaperProcessor(), # 现代研究
'expert_knowledge': ExpertKnowledgeProcessor() # 专家知识
}
def integrate_knowledge(self):
"""
整合多源知识
"""
integrated_graph = {}
for source_name, processor in self.knowledge_sources.items():
# 提取各源知识
source_knowledge = processor.extract_knowledge()
# 知识对齐
aligned_knowledge = self.align_knowledge(source_knowledge)
# 融合到总图谱
integrated_graph = self.merge_knowledge(
integrated_graph,
aligned_knowledge,
source_weight=self.get_source_weight(source_name)
)
return integrated_graph
def resolve_conflicts(self, conflicting_facts):
"""
解决知识冲突
"""
resolved_facts = []
for fact_group in conflicting_facts:
# 基于来源可信度排序
sorted_facts = sorted(
fact_group,
key=lambda x: x['source_credibility'],
reverse=True
)
# 选择最可信的事实
primary_fact = sorted_facts[0]
# 如果有多个高可信度事实,进行投票
if len([f for f in sorted_facts if f['source_credibility'] > 0.8]) > 1:
voted_fact = self.vote_on_facts(sorted_facts)
resolved_facts.append(voted_fact)
else:
resolved_facts.append(primary_fact)
return resolved_facts知识质量验证
一致性检查:
python
class KnowledgeValidator:
def __init__(self):
self.consistency_rules = [
{
'rule': 'symptom_disease_consistency',
'description': '症状与疾病的一致性检查'
},
{
'rule': 'treatment_syndrome_consistency',
'description': '治法与证候的一致性检查'
}
]
def validate_consistency(self, knowledge_graph):
"""
验证知识图谱一致性
"""
validation_results = []
for rule in self.consistency_rules:
result = self.apply_consistency_rule(knowledge_graph, rule)
validation_results.append(result)
return validation_results
def check_logical_consistency(self, entity1, relation, entity2):
"""
检查三元组的逻辑一致性
"""
# 检查实体类型匹配
if not self.check_type_compatibility(entity1, relation, entity2):
return False, "实体类型不匹配"
# 检查医学逻辑
if not self.check_medical_logic(entity1, relation, entity2):
return False, "违反医学逻辑"
return True, "通过一致性检查"图谱存储与查询
图数据库设计
Neo4j存储方案:
python
class Neo4jKnowledgeStore:
def __init__(self, uri, user, password):
self.driver = GraphDatabase.driver(uri, auth=(user, password))
def create_entity(self, entity_type, properties):
"""
创建实体节点
"""
with self.driver.session() as session:
query = f"""
CREATE (e:{entity_type} $properties)
RETURN e
"""
result = session.run(query, properties=properties)
return result.single()
def create_relationship(self, entity1_id, entity2_id, relation_type, properties=None):
"""
创建关系
"""
with self.driver.session() as session:
query = """
MATCH (e1), (e2)
WHERE id(e1) = $entity1_id AND id(e2) = $entity2_id
CREATE (e1)-[r:$relation_type $properties]->(e2)
RETURN r
"""
result = session.run(
query.replace('$relation_type', relation_type),
entity1_id=entity1_id,
entity2_id=entity2_id,
properties=properties or {}
)
return result.single()智能查询接口
图谱查询引擎:
python
class KnowledgeQueryEngine:
def __init__(self, knowledge_store):
self.store = knowledge_store
self.query_templates = {
'find_symptoms': """
MATCH (d:Disease)-[:has_symptom]->(s:Symptom)
WHERE d.name = $disease_name
RETURN s.name, s.description
""",
'find_treatments': """
MATCH (syn:Syndrome)-[:treated_by]->(t:Treatment)
WHERE syn.name = $syndrome_name
RETURN t.name, t.method, t.formula
""",
'find_similar_cases': """
MATCH (c1:Case)-[:has_symptom]->(s:Symptom)<-[:has_symptom]-(c2:Case)
WHERE c1.id = $case_id AND c1 <> c2
RETURN c2, count(s) as shared_symptoms
ORDER BY shared_symptoms DESC
LIMIT 10
"""
}
def query_related_symptoms(self, disease_name):
"""
查询疾病相关症状
"""
return self.store.execute_query(
self.query_templates['find_symptoms'],
disease_name=disease_name
)
def query_treatment_options(self, syndrome_name):
"""
查询治疗方案
"""
return self.store.execute_query(
self.query_templates['find_treatments'],
syndrome_name=syndrome_name
)图谱应用集成
智能推理增强
基于图谱的推理:
python
class GraphReasoningEngine:
def __init__(self, knowledge_graph, llm_model):
self.kg = knowledge_graph
self.llm = llm_model
def enhanced_diagnosis(self, symptoms, patient_info):
"""
基于知识图谱增强的诊断
"""
# 从图谱中查询相关信息
related_diseases = self.kg.query_diseases_by_symptoms(symptoms)
related_syndromes = self.kg.query_syndromes_by_symptoms(symptoms)
# 构建增强的上下文
enhanced_context = self.build_enhanced_context(
symptoms,
related_diseases,
related_syndromes,
patient_info
)
# LLM推理
diagnosis = self.llm.generate_diagnosis(enhanced_context)
# 图谱验证
validated_diagnosis = self.validate_with_kg(diagnosis)
return validated_diagnosis
def build_enhanced_context(self, symptoms, diseases, syndromes, patient_info):
"""
构建增强的诊断上下文
"""
context = f"患者症状:{', '.join(symptoms)}\n"
context += f"相关疾病:{', '.join([d['name'] for d in diseases])}\n"
context += f"可能证候:{', '.join([s['name'] for s in syndromes])}\n"
context += f"患者信息:{patient_info}\n"
return context知识图谱可视化
图谱可视化接口:
python
class KnowledgeGraphVisualizer:
def __init__(self, knowledge_graph):
self.kg = knowledge_graph
def generate_subgraph(self, center_entity, depth=2):
"""
生成以某实体为中心的子图
"""
subgraph_data = {
'nodes': [],
'edges': []
}
# 获取中心节点
center_node = self.kg.get_entity(center_entity)
subgraph_data['nodes'].append(self.format_node(center_node))
# 递归获取相邻节点
visited = {center_entity}
queue = [(center_entity, 0)]
while queue:
current_entity, current_depth = queue.pop(0)
if current_depth < depth:
neighbors = self.kg.get_neighbors(current_entity)
for neighbor in neighbors:
if neighbor['entity'] not in visited:
visited.add(neighbor['entity'])
subgraph_data['nodes'].append(
self.format_node(neighbor)
)
queue.append((neighbor['entity'], current_depth + 1))
# 添加边
subgraph_data['edges'].append({
'source': current_entity,
'target': neighbor['entity'],
'relation': neighbor['relation'],
'weight': neighbor.get('weight', 1.0)
})
return subgraph_data评估与优化
图谱质量评估
质量指标体系:
python
class KnowledgeGraphEvaluator:
def __init__(self, knowledge_graph):
self.kg = knowledge_graph
def evaluate_completeness(self):
"""
评估知识完整性
"""
# 计算实体覆盖率
domain_entities = self.get_domain_entities()
kg_entities = self.kg.get_all_entities()
entity_coverage = len(kg_entities) / len(domain_entities)
# 计算关系覆盖率
expected_relations = self.get_expected_relations()
actual_relations = self.kg.get_all_relations()
relation_coverage = len(actual_relations) / len(expected_relations)
return {
'entity_coverage': entity_coverage,
'relation_coverage': relation_coverage,
'overall_completeness': (entity_coverage + relation_coverage) / 2
}
def evaluate_accuracy(self, gold_standard):
"""
评估知识准确性
"""
correct_facts = 0
total_facts = 0
for fact in self.kg.get_all_facts():
total_facts += 1
if self.verify_fact_accuracy(fact, gold_standard):
correct_facts += 1
accuracy = correct_facts / total_facts if total_facts > 0 else 0
return {
'accuracy': accuracy,
'correct_facts': correct_facts,
'total_facts': total_facts
}构建优势
- 结构化知识:将非结构化的中医知识转化为可计算的结构
- 推理增强:为AI模型提供额外的知识支撑
- 知识传承:数字化保存和传播中医传统知识
- 可扩展性:支持知识的持续积累和更新
发展规划:虽然当前主要依赖GLM-4.1V的内在知识,但知识图谱的构建为未来系统升级和知识增强奠定了坚实基础。